Protein Name: 14-3-3 protein epsilon
|
UniprotKB/SwissProt ID: 1433E_MOUSE (P62259)
Gene Name: Ywhae
Organism: Mus musculus (Mouse).
Function: Adapter protein implicated in the regulation of a large spectrum of both general and specialized signaling pathways. Binds to a large number of partners, usually by recognition of a phosphoserine or phosphothreonine motif. Binding generally results in the modulation of the activity of the binding partner.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm (By similarity). Melanosome (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 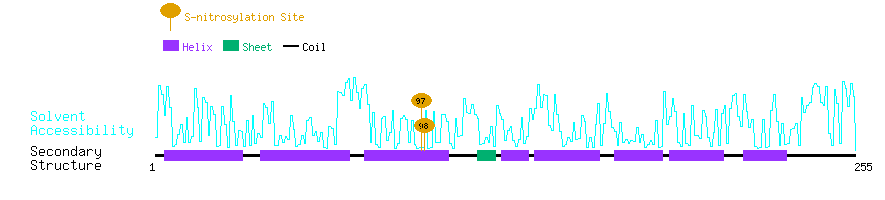 |
|

