Protein Name: Aspartate aminotransferase, mitochondrial
|
UniprotKB/SwissProt ID: AATM_MOUSE (P05202)
Gene Name: Got2
Synonyms: Got-2
Organism: Mus musculus (Mouse).
Function: Catalyzes the irreversible transamination of the L- tryptophan metabolite L-kynurenine to form kynurenic acid (KA). Plays a key role in amino acid metabolism. Important for metabolite exchange between mitochondria and cytosol. Facilitates cellular uptake of long-chain free fatty acids.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Mitochondrion matrix. Cell membrane.
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 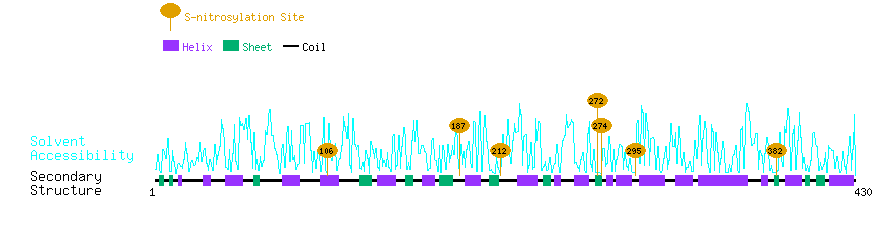 |
|
