Protein Name: Short/branched chain specific acyl-CoA dehydrogenase, mitochondrial
|
UniprotKB/SwissProt ID: ACDSB_MOUSE (Q9DBL1)
Gene Name: Acadsb
Organism: Mus musculus (Mouse).
Function: Has greatest activity toward short branched chain acyl- CoA derivative such as (s)-2-methylbutyryl-CoA, isobutyryl-CoA, and 2-methylhexanoyl-CoA as well as toward short straight chain acyl-CoAs such as butyryl-CoA and hexanoyl-CoA. Can use valproyl- CoA as substrate and may play a role in controlling the metabolic flux of valproic acid in the development of toxicity of this agent (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Mitochondrion matrix (Probable).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 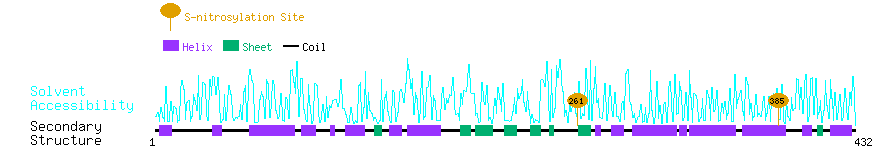 |
|

