Protein Name: Acetyl-coenzyme A synthetase 2-like, mitochondrial
|
UniprotKB/SwissProt ID: ACS2L_MOUSE (Q99NB1)
Gene Name: Acss1
Synonyms: Acas2, Acas2l
Organism: Mus musculus (Mouse).
Function: Important for maintaining normal body temperature during fasting and for energy homeostasis. Essential for energy expenditure under ketogenic conditions. Converts acetate to acetyl-CoA so that it can be used for oxidation through the tricarboxylic cycle to produce ATP and CO(2).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Mitochondrion matrix.
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 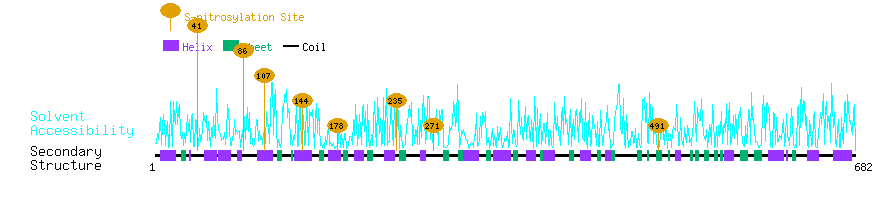 |
|

