Protein Name: Long-chain-fatty-acid--CoA ligase 1
|
UniprotKB/SwissProt ID: ACSL1_MOUSE (P41216)
Gene Name: Acsl1
Synonyms: Acsl2, Facl2
Organism: Mus musculus (Mouse).
Function: Activation of long-chain fatty acids for both synthesis of cellular lipids, and degradation via beta-oxidation. Preferentially uses oleate, arachidonate, eicosapentaenoate and docosahexaenoate as substrates (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Mitochondrion outer membrane; Single-pass type III membrane protein (By similarity). Peroxisome membrane; Single-pass type III membrane protein (By similarity). Microsome membrane; Single-pass type III membrane protein (By similarity). Endoplasmic reticu
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 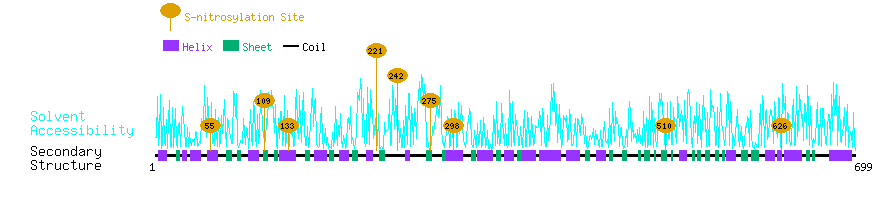 |
|

