Protein Name: Long-chain-fatty-acid--CoA ligase 5
|
UniprotKB/SwissProt ID: ACSL5_MOUSE (Q8JZR0)
Gene Name: Acsl5
Synonyms: Facl5
Organism: Mus musculus (Mouse).
Function: Acyl-CoA synthetases (ACSL) activates long-chain fatty acids for both synthesis of cellular lipids, and degradation via beta-oxidation. ACSL5 may activate fatty acids from exogenous sources for the synthesis of triacylglycerol destined for intracellular storage (By similarity). It was suggested that it may also stimulate fatty acid oxidation (By similarity). At the villus tip of the crypt-villus axis of the small intestine may sensitize epithelial cells to apoptosis specifically triggered by the death ligand TRAIL (By similarity). May have a role in the survival of glioma cells (By similarity). Utilizes a wide range of saturated fatty acids with a preference for C16-C18 unsaturated fatty acids (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Mitochondrion (By similarity). Endoplasmic reticulum (By similarity). Mitochondrion outer membrane; Single- pass type III membrane protein (By similarity). Endoplasmic reticulum membrane; Single-pass type III membrane protein (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 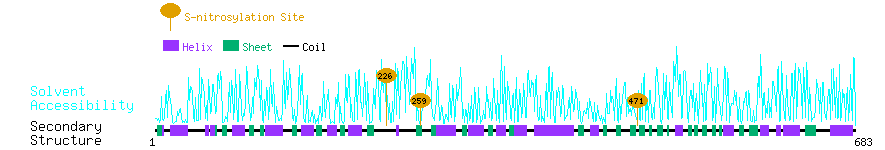 |
|

