Protein Name: Alpha-actinin-4
|
UniprotKB/SwissProt ID: ACTN4_MOUSE (P57780)
Gene Name: Actn4
Organism: Mus musculus (Mouse).
Function: F-actin cross-linking protein which is thought to anchor actin to a variety of intracellular structures. This is a bundling protein. Probably involved in vesicular trafficking via its association with the CART complex. The CART complex is necessary for efficient transferrin receptor recycling but not for EGFR degradation. Involved in tight junction assembly in epithelial cells probably through interaction with MICALL2. Links MICALL2 to the actin cytoskeleton and recruits it to the tight junctions.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Nucleus (By similarity). Cytoplasm (By similarity). Cell junction. Note=Localized in cytoplasmic mRNP granules containing untranslated mRNAs. Colocalizes with actin stress fibers (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 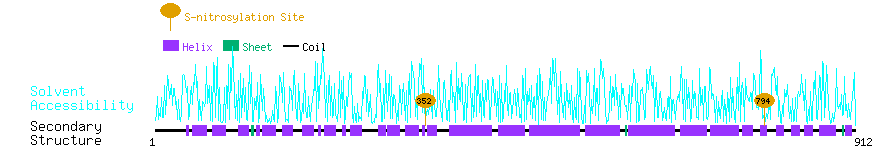 |
|

