Protein Name: N-acyl-aromatic-L-amino acid amidohydrolase (carboxylate-forming)
|
UniprotKB/SwissProt ID: ACY3_MOUSE (Q91XE4)
Gene Name: Acy3
Synonyms: Aspa2
Organism: Mus musculus (Mouse).
Function: Plays an important role in deacetylating mercapturic acids in kidney proximal tubules. Also acts on N-acetyl-aromatic amino acids.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Apical cell membrane; Peripheral membrane protein. Cytoplasm. Note=Predominantly localized in the apical membrane of cells in the S1 segment. In the proximal straight tubules (S2 and S3 segments) is expressed diffusely throughout the cytoplasm.
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 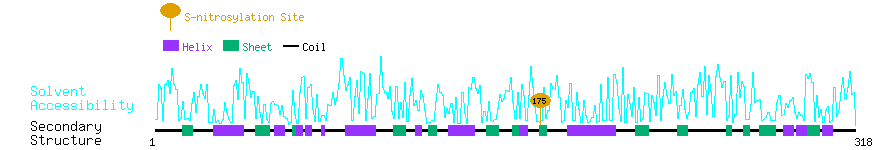 |
|
