Protein Name: Type-1 angiotensin II receptor
|
UniprotKB/SwissProt ID: AGTR1_HUMAN (P30556)
Gene Name: AGTR1
Synonyms: AGTR1A, AGTR1B, AT2R1, AT2R1B
Organism: Homo sapiens (Human).
Function: Receptor for angiotensin II. Mediates its action by association with G proteins that activate a phosphatidylinositol- calcium second messenger system.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cell membrane; Multi-pass membrane protein.
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | KEGG | H00575 | Renal tubular dysgenesis | | OMIM | 106165 | ANGIOTENSIN RECEPTOR 1; AGTR1 ;;ANGIOTENSIN II RECEPTOR, VASCULAR TYPE 1; AT2R1;; ANGIOTENSI | | OMIM | 145500 | HYPERTENSION, ESSENTIAL ;;EHT | | OMIM | 267430 | RENAL TUBULAR DYSGENESIS; RTD ;;PRIMITIVE RENAL TUBULE SYNDROME RENAL TUBULAR DYSGENESIS WIT | | HPRD | 00107 | Hypertension, essential, susceptibility to | | HPRD | 00107 | Renal tubular dysgenesis | | HPRD | 00107 | Stature as a quantitative trait |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 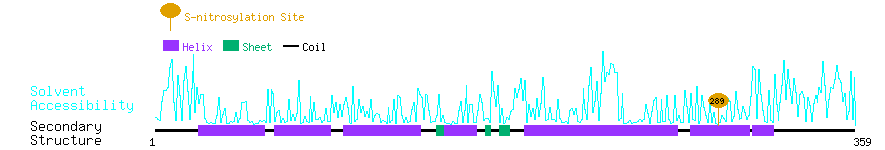 |
|
