Protein Name: Sodium/potassium-transporting ATPase subunit alpha-3
|
UniprotKB/SwissProt ID: AT1A3_MOUSE (Q6PIC6)
Gene Name: Atp1a3
Organism: Mus musculus (Mouse).
Function: This is the catalytic component of the active enzyme, which catalyzes the hydrolysis of ATP coupled with the exchange of sodium and potassium ions across the plasma membrane. This action creates the electrochemical gradient of sodium and potassium ions, providing the energy for active transport of various nutrients (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cell membrane; Multi-pass membrane protein (By similarity).
| |
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 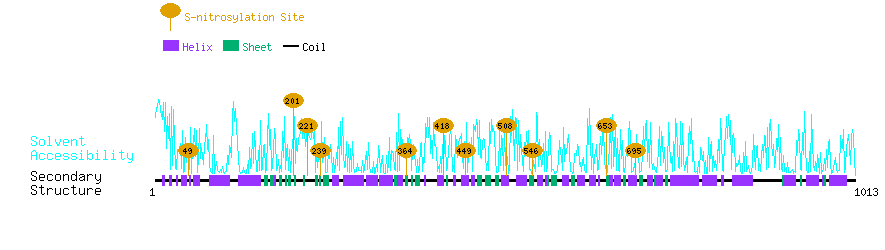 |
|

