Protein Name: ATP synthase subunit s, mitochondrial
|
UniprotKB/SwissProt ID: ATP5S_MOUSE (Q9CRA7)
Gene Name: Atp5s
Synonyms: Atpw
Organism: Mus musculus (Mouse).
Function: Involved in regulation of mitochondrial membrane ATP synthase. Necessary for H(+) conduction of ATP synthase. Facilitates energy-driven catalysis of ATP synthesis by blocking a proton leak through an alternative proton exit pathway.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Mitochondrion (By similarity). Mitochondrion inner membrane (By similarity).
| |
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 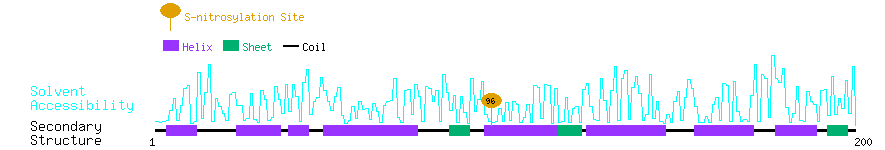 |
|

