Protein Name: ATP synthase subunit alpha, mitochondrial
|
UniprotKB/SwissProt ID: ATPA_MOUSE (Q03265)
Gene Name: Atp5a1
Organism: Mus musculus (Mouse).
Function: Mitochondrial membrane ATP synthase (F(1)F(0) ATP synthase or Complex V) produces ATP from ADP in the presence of a proton gradient across the membrane which is generated by electron transport complexes of the respiratory chain. F-type ATPases consist of two structural domains, F(1) - containing the extramembraneous catalytic core, and F(0) - containing the membrane proton channel, linked together by a central stalk and a peripheral stalk. During catalysis, ATP synthesis in the catalytic domain of F(1) is coupled via a rotary mechanism of the central stalk subunits to proton translocation. Subunits alpha and beta form the catalytic core in F(1). Rotation of the central stalk against the surrounding alpha(3)beta(3) subunits leads to hydrolysis of ATP in three separate catalytic sites on the beta subunits. Subunit alpha does not bear the catalytic high-affinity ATP-binding sites (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Mitochondrion inner membrane (By similarity). Cell membrane; Peripheral membrane protein; Extracellular side (By similarity). Note=Colocalizes with HRG on the cell surface of T-cells (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 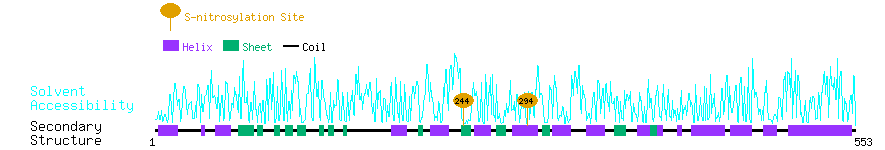 |
|

