Protein Name: Methylglutaconyl-CoA hydratase, mitochondrial
|
UniprotKB/SwissProt ID: AUHM_MOUSE (Q9JLZ3)
Gene Name: Auh
Organism: Mus musculus (Mouse).
Function: Catalyzes the conversion of 3-methylglutaconyl-CoA to 3- hydroxy-3-methylglutaryl-CoA. Has very low enoyl-CoA hydratase activity. Was originally identified as RNA-binding protein that binds in vitro to clustered 5'-AUUUA-3' motifs (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Mitochondrion.
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 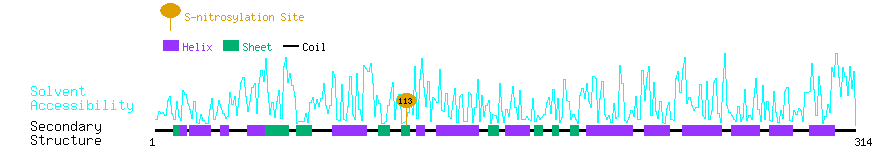 |
|

