Protein Name: Apoptosis regulator Bcl-2
|
UniprotKB/SwissProt ID: BCL2_HUMAN (P10415)
Gene Name: BCL2
Organism: Homo sapiens (Human).
Function: Suppresses apoptosis in a variety of cell systems including factor-dependent lymphohematopoietic and neural cells. Regulates cell death by controlling the mitochondrial membrane permeability. Appears to function in a feedback loop system with caspases. Inhibits caspase activity either by preventing the release of cytochrome c from the mitochondria and/or by binding to the apoptosis-activating factor (APAF-1).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Mitochondrion outer membrane; Single-pass membrane protein. Nucleus membrane; Single-pass membrane protein. Endoplasmic reticulum membrane; Single-pass membrane protein.
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | KEGG | H00005 | Chronic lymphocytic leukemia (CLL) | | KEGG | H00013 | Small cell lung cancer | | KEGG | H00018 | Gastric cancer | | KEGG | H00028 | Choriocarcinoma | | KEGG | H00030 | Cervical cancer | | KEGG | H00041 | Kaposi's sarcoma | | KEGG | H00054 | Nasopharyngeal cancer | | KEGG | H00020 | Colorectal cancer | | OMIM | 151430 | B-CELL CLL/LYMPHOMA 2; BCL2 ;;ONCOGENE B-CELL LEUKEMIA 2 LEUKEMIA/LYMPHOMA, B-CELL, 2 |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 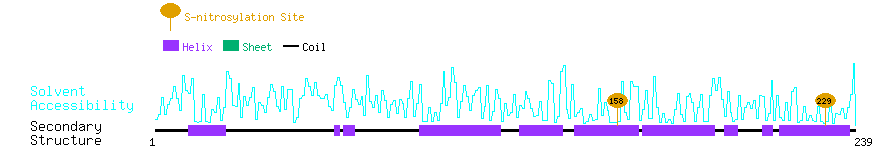 |
|
