Protein Name: Calpain-2 catalytic subunit
|
UniprotKB/SwissProt ID: CAN2_MOUSE (O08529)
Gene Name: Capn2
Organism: Mus musculus (Mouse).
Function: Calcium-regulated non-lysosomal thiol-protease which catalyze limited proteolysis of substrates involved in cytoskeletal remodeling and signal transduction (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm (By similarity). Cell membrane (By similarity). Note=Translocates to the plasma membrane upon Ca(2+) binding (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 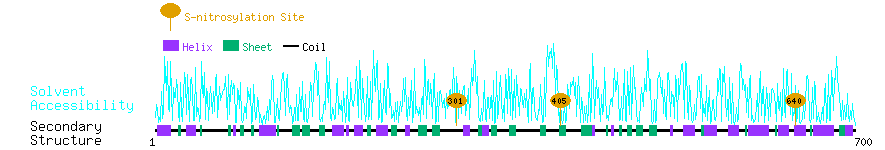 |
|

