Protein Name: Cathepsin K
|
UniprotKB/SwissProt ID: CATK_HUMAN (P43235)
Gene Name: CTSK
Synonyms: CTSO, CTSO2
Organism: Homo sapiens (Human).
Function: Closely involved in osteoclastic bone resorption and may participate partially in the disorder of bone remodeling. Displays potent endoprotease activity against fibrinogen at acid pH. May play an important role in extracellular matrix degradation.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Lysosome.
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | HPRD | 03064 | Pycnodysostosis |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 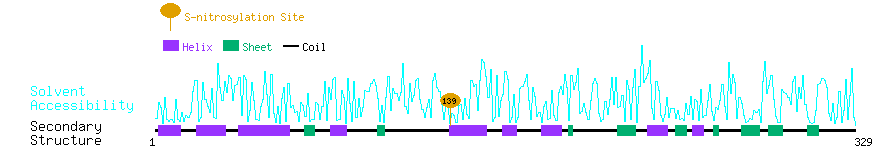 |
|
