Protein Name: Caveolin-3
|
UniprotKB/SwissProt ID: CAV3_MOUSE (P51637)
Gene Name: Cav3
Organism: Mus musculus (Mouse).
Function: May act as a scaffolding protein within caveolar membranes. Interacts directly with G-protein alpha subunits and can functionally regulate their activity. May also regulate voltage-gated potassium channels. Plays a role in the sarcolemma repair mechanism of both skeletal muscle and cardiomyocytes that permits rapid resealing of membranes disrupted by mechanical stress.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Golgi apparatus membrane; Peripheral membrane protein (By similarity). Cell membrane; Peripheral membrane protein (By similarity). Membrane, caveola; Peripheral membrane protein (By similarity). Note=Potential hairpin-like structure in the membrane. Memb
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 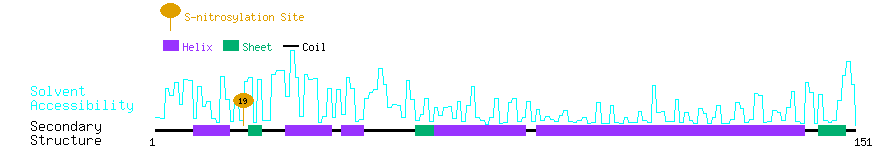 |
|

