Protein Name: 60 kDa heat shock protein, mitochondrial
|
UniprotKB/SwissProt ID: CH60_MOUSE (P63038)
Gene Name: Hspd1
Synonyms: Hsp60
Organism: Mus musculus (Mouse).
Function: Implicated in mitochondrial protein import and macromolecular assembly. May facilitate the correct folding of imported proteins. May also prevent misfolding and promote the refolding and proper assembly of unfolded polypeptides generated under stress conditions in the mitochondrial matrix.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Mitochondrion matrix (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 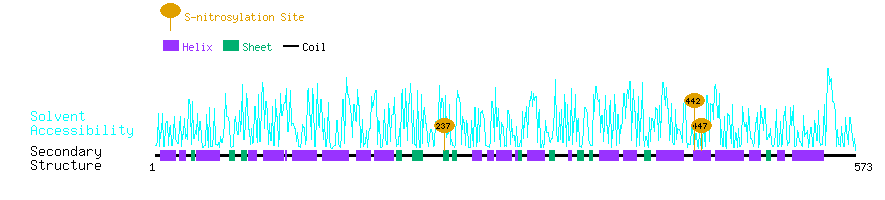 |
|

