Protein Name: L-xylulose reductase
|
UniprotKB/SwissProt ID: DCXR_MOUSE (Q91X52)
Gene Name: Dcxr
Organism: Mus musculus (Mouse).
Function: Catalyzes the NADPH-dependent reduction of several pentoses, tetroses, trioses, alpha-dicarbonyl compounds and L- xylulose. Participates in the uronate cycle of glucose metabolism. May play a role in the water absorption and cellular osmoregulation in the proximal renal tubules by producing xylitol, an osmolyte, thereby preventing osmolytic stress from occurring in the renal tubules.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Membrane; Peripheral membrane protein (By similarity). Apical cell membrane; Peripheral membrane protein. Note=Probably recruited to membranes via an interaction with phosphatidylinositol (By similarity). In kidney, it is localized in the brush border me
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 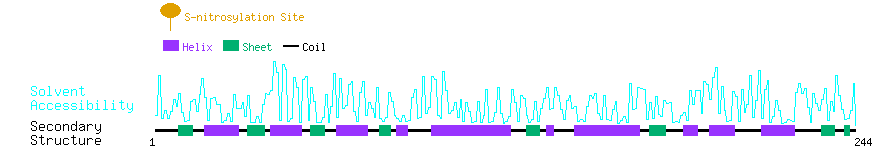 |
|

