Protein Name: Probable ATP-dependent RNA helicase DDX17
|
UniprotKB/SwissProt ID: DDX17_MOUSE (Q501J6)
Gene Name: Ddx17
Organism: Mus musculus (Mouse).
Function: RNA-dependent ATPase activity. Involved in transcriptional regulation. Transcriptional coactivator for estrogen receptor ESR1. Increases ESR1 AF-1 domain-mediated transactivation (By similarity). Synergizes with DDX5 and SRA1 RNA to activate MYOD1 transcriptional activity and probably involved in skeletal muscle differentiation. Required for zinc-finger antiviral protein ZC3HAV1-mediated mRNA degradation.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Nucleus (By similarity). Nucleus, nucleolus (By similarity).
| |
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 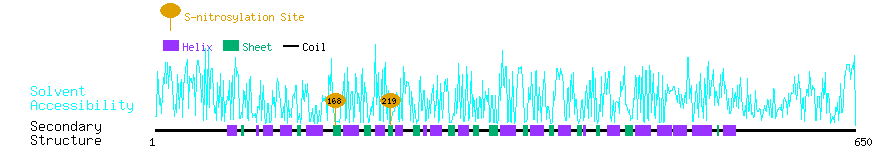 |
|

