Protein Name: Peroxisomal multifunctional enzyme type 2
|
UniprotKB/SwissProt ID: DHB4_MOUSE (P51660)
Gene Name: Hsd17b4
Synonyms: Edh17b4
Organism: Mus musculus (Mouse).
Function: Bifunctional enzyme acting on the peroxisomal beta- oxidation pathway for fatty acids. Catalyzes the formation of 3- ketoacyl-CoA intermediates from both straight-chain and 2-methyl- branched-chain fatty acids (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Peroxisome (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 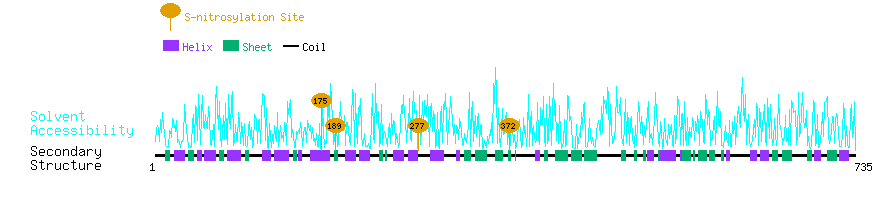 |
|

