Protein Name: Glutamate dehydrogenase 1, mitochondrial
|
UniprotKB/SwissProt ID: DHE3_MOUSE (P26443)
Gene Name: Glud1
Synonyms: Glud
Organism: Mus musculus (Mouse).
Function: Mitochondrial glutamate dehydrogenase that converts L- glutamate into alpha-ketoglutarate. Plays a key role in glutamine anaplerosis by producing alpha-ketoglutarate, an important intermediate in the tricarboxylic acid cycle. May be involved in learning and memory reactions by increasing the turnover of the excitatory neurotransmitter glutamate.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Mitochondrion matrix.
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 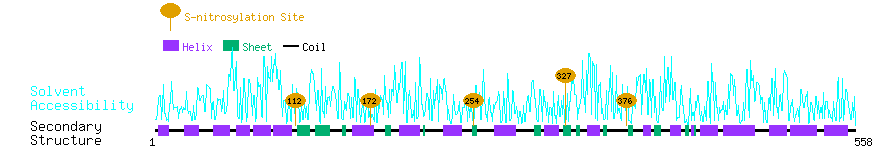 |
|

