Protein Name: Protein diaphanous homolog 1
|
UniprotKB/SwissProt ID: DIAP1_HUMAN (O60610)
Gene Name: DIAPH1
Synonyms: DIAP1
Organism: Homo sapiens (Human).
Function: Acts in a Rho-dependent manner to recruit PFY1 to the membrane. Required for the assembly of F-actin structures, such as actin cables and stress fibers. Nucleates actin filaments. Binds to the barbed end of the actin filament and slows down actin polymerization and depolymerization. Required for cytokinesis, and transcriptional activation of the serum response factor. DFR proteins couple Rho and Src tyrosine kinase during signaling and the regulation of actin dynamics. Functions as a scaffold protein for MAPRE1 and APC to stabilize microtubules and promote cell migration (By similarity). Has neurite outgrowth promoting activity (By similarity). In hear cells, it may play a role in the regulation of actin polymerization in hair cells. The MEMO1-RHOA- DIAPH1 signaling pathway plays an important role in ERBB2- dependent stabilization of microtubules at the cell cortex. It controls the localization of APC and CLASP2 to the cell membrane, via the regulation of GSK3B activity. In turn, membrane-bound APC allows the localization of the MACF1 to the cell membrane, which is required for microtubule capture and stabilization. Plays a role in the regulation of cell morphology and cytoskeletal organization. Required in the control of cell shape.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cell membrane. Cell projection, ruffle membrane. Cytoplasm, cytoskeleton. Note=Membrane ruffles, especially at the tip of ruffles, of motile cells.
| |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | KEGG | H00604 | Deafness, autosomal dominant | | OMIM | 124900 | DEAFNESS, AUTOSOMAL DOMINANT 1; DFNA1 ;;HEREDITARY LOW FREQUENCY HEARING LOSS; LFHL1;; DEAFN | | OMIM | 602121 | DIAPHANOUS, DROSOPHILA, HOMOLOG OF, 1; DIAPH1 ;;DIA1 | | HPRD | 03670 | Deafness, autosomal dominant nonsyndromic sensorineural, 1; DFNA1 |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 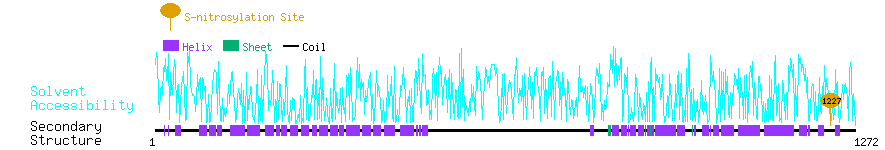 |
|

