Protein Name: Dihydropyrimidinase-related protein 2
|
UniprotKB/SwissProt ID: DPYL2_MOUSE (O08553)
Gene Name: Dpysl2
Synonyms: Crmp2, Ulip2
Organism: Mus musculus (Mouse).
Function: Plays a role in neuronal development and polarity, as well as in axon growth and guidance, neuronal growth cone collapse and cell migration. Necessary for signaling by class 3 semaphorins and subsequent remodeling of the cytoskeleton. May play a role in endocytosis.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm, cytosol (By similarity). Cytoplasm, cytoskeleton (By similarity). Membrane (By similarity). Note=Tightly but non-covalently associated with membranes (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 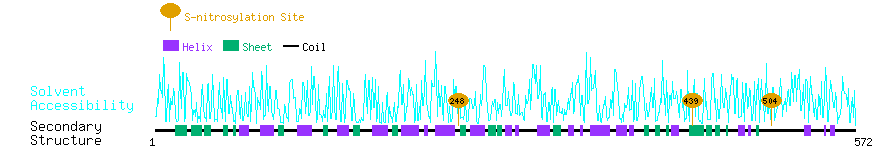 |
|

