Protein Name: Gamma-enolase
|
UniprotKB/SwissProt ID: ENOG_RAT (P07323)
Gene Name: Eno2
Synonyms: Eno-2
Organism: Rattus norvegicus (Rat).
Function: Has neurotrophic and neuroprotective properties on a broad spectrum of central nervous system (CNS) neurons. Binds, in a calcium-dependent manner, to cultured neocortical neurons and promotes cell survival.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm (By similarity). Cell membrane (By similarity). Note=Can translocate to the plasma membrane in either the homodimeric (alpha/alpha) or heterodimeric (alpha/gamma) form (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 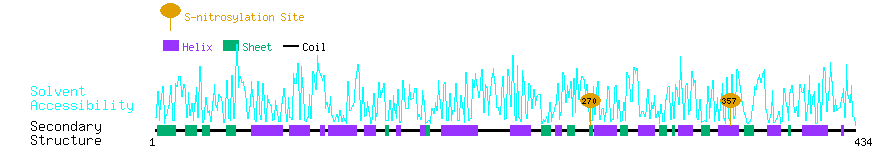 |
|

