Protein Name: Eyes absent homolog 4
|
UniprotKB/SwissProt ID: EYA4_HUMAN (O95677)
Gene Name: EYA4
Organism: Homo sapiens (Human).
Function: Tyrosine phosphatase that specifically dephosphorylates 'Tyr-142' of histone H2AX (H2AXY142ph). 'Tyr-142' phosphorylation of histone H2AX plays a central role in DNA repair and acts as a mark that distinguishes between apoptotic and repair responses to genotoxic stress. Promotes efficient DNA repair by dephosphorylating H2AX, promoting the recruitment of DNA repair complexes containing MDC1. Its function as histone phosphatase probably explains its role in transcription regulation during organogenesis. May be involved in development of the eye (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm (By similarity). Nucleus (By similarity).
| |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | HPRD | 04648 | Cardiomyopathy, dilated, 1j | | HPRD | 04648 | Deafness, autosomal dominant nonsyndromic sensorineural 10 |
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 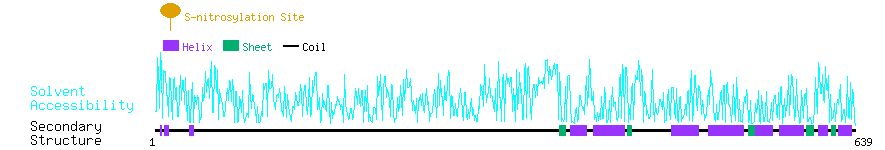 |
|

