Protein Name: Fatty acid-binding protein, epidermal
|
UniprotKB/SwissProt ID: FABP5_MOUSE (Q05816)
Gene Name: Fabp5
Synonyms: Fabpe, Klbp, Mal1
Organism: Mus musculus (Mouse).
Function: High specificity for fatty acids. Highest affinity for C18 chain length (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm.
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 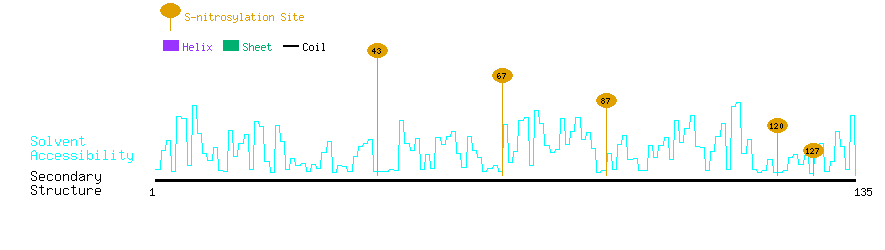 |
|
