Protein Name: F-box only protein 7
|
UniprotKB/SwissProt ID: FBX7_HUMAN (Q9Y3I1)
Gene Name: FBXO7
Synonyms: FBX7
Organism: Homo sapiens (Human).
Function: Substrate recognition component of a SCF (SKP1-CUL1-F- box protein) E3 ubiquitin-protein ligase complex which mediates the ubiquitination and subsequent proteasomal degradation of target proteins. Recognizes BIRC2 and DLGAP5. Plays a role downstream of PINK1 in the clearance of damaged mitochondria via selective autophagy (mitophagy) by targeting PARK2 to dysfunctional depolarized mitochondria. Promotes MFN1 ubiquitination.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm. Nucleus. Mitochondrion. Cytoplasm, cytosol. Note=Predominantly cytoplasmic. A minor proportion is detected in the nucleus. Relocates from the cytosol to depolarized mitochondria.
| |
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 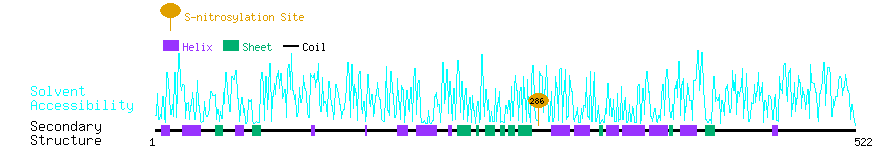 |
|
