Protein Name: Glyceraldehyde-3-phosphate dehydrogenase
|
UniprotKB/SwissProt ID: G3P_MOUSE (P16858)
Gene Name: Gapdh
Synonyms: Gapd
Organism: Mus musculus (Mouse).
Function: Has both glyceraldehyde-3-phosphate dehydrogenase and nitrosylase activities, thereby playing a role in glycolysis and nuclear functions, respectively. Glyceraldehyde-3-phosphate dehydrogenase is a key enzyme in glycolysis that catalyzes the first step of the pathway by converting D-glyceraldehyde 3- phosphate (G3P) into 3-phospho-D-glyceroyl phosphate. Modulates the organization and assembly of the cytoskeleton. Facilitates the CHP1-dependent microtubule and membrane associations through its ability to stimulate the binding of CHP1 to microtubules. Also participates in nuclear events including transcription, RNA transport, DNA replication and apoptosis. Nuclear functions are probably due to the nitrosylase activity that mediates cysteine S- nitrosylation of nuclear target proteins such as SIRT1, HDAC2 and PRKDC. Component of the GAIT (gamma interferon-activated inhibitor of translation) complex which mediates interferon-gamma-induced transcript-selective translation inhibition in inflammation processes. Upon interferon-gamma treatment assembles into the GAIT complex which binds to stem loop-containing GAIT elements in the 3'-UTR of diverse inflammatory mRNAs (such as ceruplasmin) and suppresses their translation.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm, cytosol (By similarity). Nucleus (By similarity). Cytoplasm, cytoskeleton (By similarity). Note=Translocates to the nucleus following S-nitrosylation and interaction with SIAH1, which contains a nuclear localization signal (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 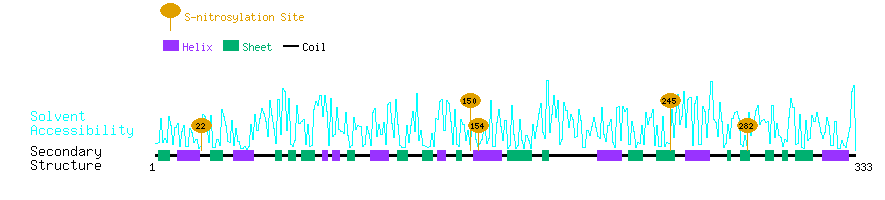 |
|

