Protein Name: Glutaryl-CoA dehydrogenase, mitochondrial
|
UniprotKB/SwissProt ID: GCDH_MOUSE (Q60759)
Gene Name: Gcdh
Organism: Mus musculus (Mouse).
Function: Catalyzes the oxidative decarboxylation of glutaryl-CoA to crotonyl-CoA and CO(2) in the degradative pathway of L-lysine, L-hydroxylysine, and L-tryptophan metabolism. It uses electron transfer flavoprotein as its electron acceptor.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Mitochondrion matrix.
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 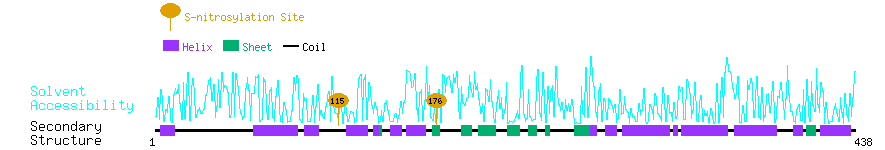 |
|

