Protein Name: Glutamine synthetase
|
UniprotKB/SwissProt ID: GLNA_MOUSE (P15105)
Gene Name: Glul
Synonyms: Glns
Organism: Mus musculus (Mouse).
Function: Essential for proliferation of fetal skin fibroblasts. This enzyme has 2 functions: it catalyzes the production of glutamine and 4-aminobutanoate (gamma-aminobutyric acid, GABA), the latter in a pyridoxal phosphate-independent manner (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm. Mitochondrion (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 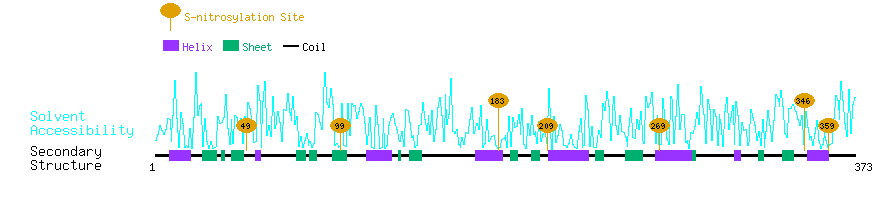 |
|

