Protein Name: Glutathione S-transferase Mu 7
|
UniprotKB/SwissProt ID: GSTM7_MOUSE (Q80W21)
Gene Name: Gstm7
Organism: Mus musculus (Mouse).
Function: Conjugation of reduced glutathione to a wide number of exogenous and endogenous hydrophobic electrophiles (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm (By similarity).
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 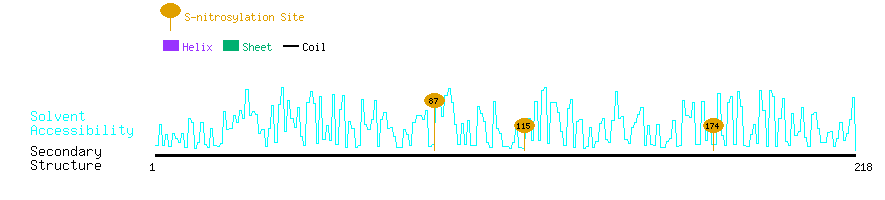 |
|
