Protein Name: Glutathione S-transferase P
|
UniprotKB/SwissProt ID: GSTP1_RAT (P04906)
Gene Name: Gstp1
Organism: Rattus norvegicus (Rat).
Function: Conjugation of reduced glutathione to a wide number of exogenous and endogenous hydrophobic electrophiles. Regulates negatively CDK5 activity via p25/p35 translocation to prevent neurodegeneration (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm (By similarity). Mitochondrion (By similarity). Nucleus (By similarity). Note=The 83 N-terminal amino acids function as un uncleaved transit peptide, and arginine residues within it are crucial for mitochondrial localization (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 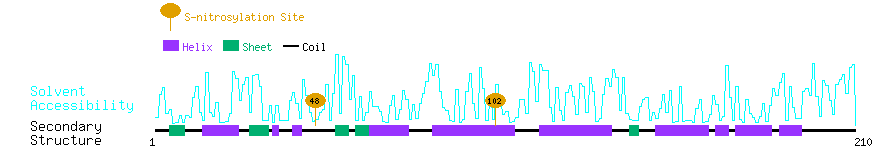 |
|

