Protein Name: Histone acetyltransferase type B catalytic subunit
|
UniprotKB/SwissProt ID: HAT1_MOUSE (Q8BY71)
Gene Name: Hat1
Organism: Mus musculus (Mouse).
Function: Acetylates soluble but not nucleosomal histone H4 at 'Lys-5' (H4K5ac) and 'Lys-12' (H4K12ac) and, to a lesser extent, acetylates histone H2A at 'Lys-5' (H2AK5ac). Has intrinsic substrate specificity that modifies lysine in recognition sequence GXGKXG. May be involved in nucleosome assembly during DNA replication and repair as part of the histone H3.1 and H3.3 complexes (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Nucleus matrix (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 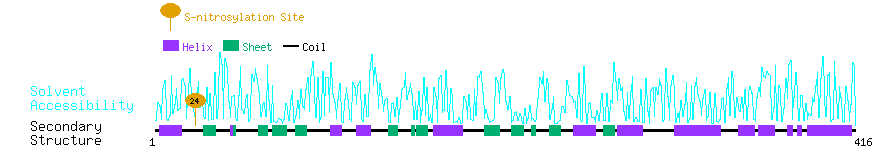 |
|

