Protein Name: Host cell factor 1
|
UniprotKB/SwissProt ID: HCFC1_HUMAN (P51610)
Gene Name: HCFC1
Synonyms: HCF1, HFC1
Organism: Homo sapiens (Human).
Function: Involved in control of the cell cycle. Also antagonizes transactivation by ZBTB17 and GABP2; represses ZBTB17 activation of the p15(INK4b) promoter and inhibits its ability to recruit p300. Coactivator for EGR2 and GABP2. Tethers the chromatin modifying Set1/Ash2 histone H3 'Lys-4' methyltransferase (H3K4me) and Sin3 histone deacetylase (HDAC) complexes (involved in the activation and repression of transcription, respectively) together. Component of a THAP1/THAP3-HCFC1-OGT complex that is required for the regulation of the transcriptional activity of RRM1. As part of the NSL complex it may be involved in acetylation of nucleosomal histone H4 on several lysine residues. In case of human herpes simplex virus (HSV) infection, HCFC1 forms a multiprotein-DNA complex with the viral transactivator protein VP16 and POU2F1 thereby enabling the transcription of the viral immediate early genes.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm. Nucleus. Note=HCFC1R1 modulates its subcellular localization and overexpression of HCFC1R1 leads to accumulation of HCFC1 in the cytoplasm. Nuclear in general, but uniquely cytoplasmic in trigeminal ganglia, becoming nuclear upon HSV reactivat
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 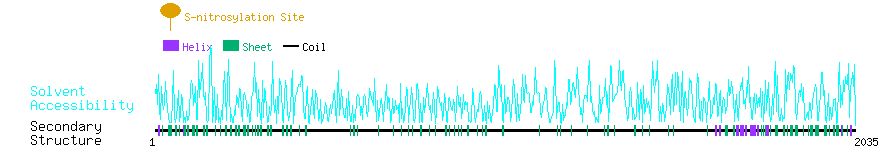 |
|

