Protein Name: Heat shock protein 105 kDa
|
UniprotKB/SwissProt ID: HS105_RAT (Q66HA8)
Gene Name: Hsph1
Synonyms: Hsp105, Hsp110
Organism: Rattus norvegicus (Rat).
Function: Prevents the aggregation of denatured proteins in cells under severe stress, on which the ATP levels decrease markedly. Inhibits HSPA8/HSC70 ATPase and chaperone activities (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 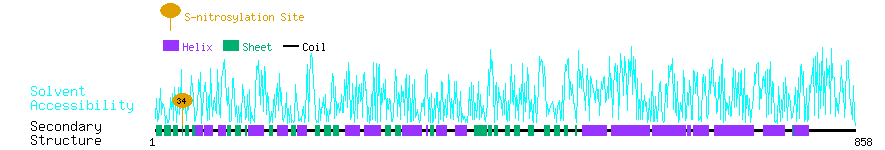 |
|

