Protein Name: Glucokinase
|
UniprotKB/SwissProt ID: HXK4_HUMAN (P35557)
Gene Name: GCK
Organism: Homo sapiens (Human).
Function: Catalyzes the initial step in utilization of glucose by the beta-cell and liver at physiological glucose concentration. Glucokinase has a high Km for glucose, and so it is effective only when glucose is abundant. The role of GCK is to provide G6P for the synthesis of glycogen. Pancreatic glucokinase plays an important role in modulating insulin secretion. Hepatic glucokinase helps to facilitate the uptake and conversion of glucose by acting as an insulin-sensitive determinant of hepatic glucose usage.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization:
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | HPRD | 00680 | Diabetes mellitus, gestational | | HPRD | 00680 | Diabetes mellitus, noninsulin-dependent, late-onset | | HPRD | 00680 | Diabetes mellitus, permanent neonatal | | HPRD | 00680 | Hyperinsulinemic hypoglycemia, familial, 3 | | HPRD | 00680 | Maturity-onset diabetes of the young, type II |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 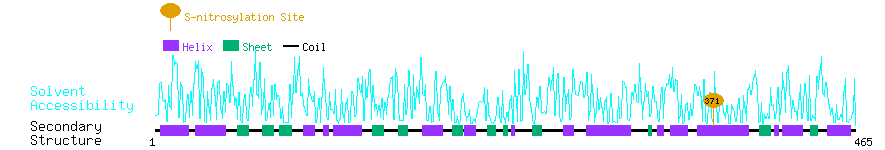 |
|
