Protein Name: Ras GTPase-activating-like protein IQGAP1
|
UniprotKB/SwissProt ID: IQGA1_HUMAN (P46940)
Gene Name: IQGAP1
Synonyms: KIAA0051
Organism: Homo sapiens (Human).
Function: Binds to activated CDC42 but does not stimulate its GTPase activity. It associates with calmodulin. Could serve as an assembly scaffold for the organization of a multimolecular complex that would interface incoming signals to the reorganization of the actin cytoskeleton at the plasma membrane. May promote neurite outgrowth.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cell membrane.
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | OMIM | 603379 | IQ MOTIF-CONTAINING GTPase-ACTIVATING PROTEIN 1; IQGAP1 ;;RASGAP-LIKE WITH IQ MOTIFS;; p195; |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 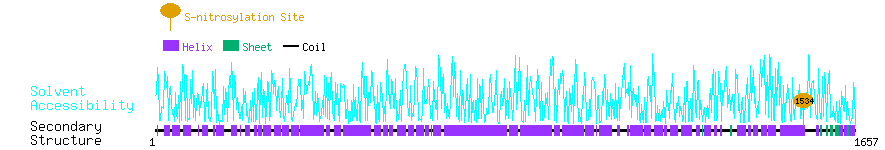 |
|

