Protein Name: Integrin alpha-6
|
UniprotKB/SwissProt ID: ITA6_HUMAN (P23229)
Gene Name: ITGA6
Organism: Homo sapiens (Human).
Function: Integrin alpha-6/beta-1 is a receptor for laminin on platelets. Integrin alpha-6/beta-4 is a receptor for laminin in epithelial cells and it plays a critical structural role in the hemidesmosome.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cell membrane; Single-pass type I membrane protein.
| |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | KEGG | H00586 | Epidermolysis bullosa, junctional, including: Epidermolysis bullosa, junctional, Herlitz type (JEB-H | | OMIM | 147556 | INTEGRIN, ALPHA-6; ITGA6 | | OMIM | 226730 | EPIDERMOLYSIS BULLOSA JUNCTIONALIS WITH PYLORIC ATRESIA ;;EPIDERMOLYSIS BULLOSA, JUNCTIONAL, | | HPRD | 00945 | Epidermolysis bullosa with pyloric atresia |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 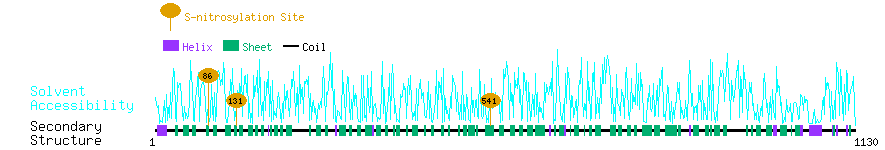 |
|

