Protein Name: Inosine triphosphate pyrophosphatase
|
UniprotKB/SwissProt ID: ITPA_MOUSE (Q9D892)
Gene Name: Itpa
Organism: Mus musculus (Mouse).
Function: Pyrophosphatase that hydrolyzes the non-canonical purine nucleotides inosine triphosphate (ITP), deoxyinosine triphosphate (dITP) as well as 2'-deoxy-N-6-hydroxylaminopurine triposphate (dHAPTP) and xanthosine 5'-triphosphate (XTP) to their respective monophosphate derivatives. The enzyme does not distinguish between the deoxy- and ribose forms. Probably excludes non-canonical purines from RNA and DNA precursor pools, thus preventing their incorporation into RNA and DNA and avoiding chromosomal lesions.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 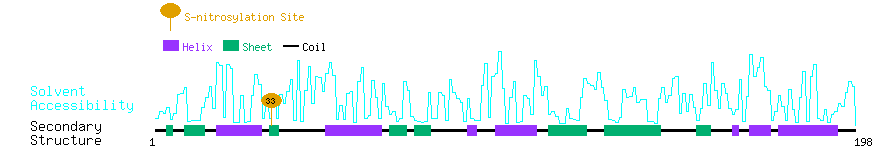 |
|

