Protein Name: Kynurenine 3-monooxygenase
|
UniprotKB/SwissProt ID: KMO_MOUSE (Q91WN4)
Gene Name: Kmo
Organism: Mus musculus (Mouse).
Function: Catalyzes the hydroxylation of L-kynurenine (L-Kyn) to form 3-hydroxy-L-kynurenine (L-3OHKyn). Required for synthesis of quinolinic acid, a neurotoxic NMDA receptor antagonist and potential endogenous inhibitor of NMDA receptor signaling in axonal targeting, synaptogenesis and apoptosis during brain development. Quinolinic acid may also affect NMDA receptor signaling in pancreatic beta cells, osteoblasts, myocardial cells, and the gastrointestinal tract (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Mitochondrion outer membrane; Multi-pass membrane protein (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 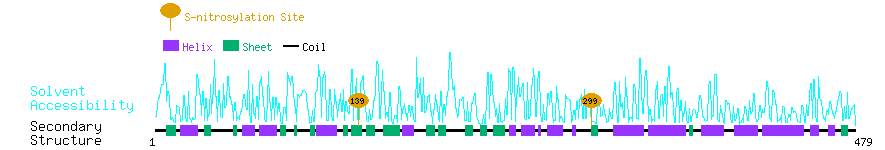 |
|

