Protein Name: Pyruvate kinase PKM
|
UniprotKB/SwissProt ID: KPYM_RAT (P11980)
Gene Name: Pkm
Synonyms: Pkm2, Pykm
Organism: Rattus norvegicus (Rat).
Function: Glycolytic enzyme that catalyzes the transfer of a phosphoryl group from phosphoenolpyruvate (PEP) to ADP, generating ATP. Stimulates POU5F1-mediated transcriptional activation (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cytoplasm (By similarity). Nucleus (By similarity).
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 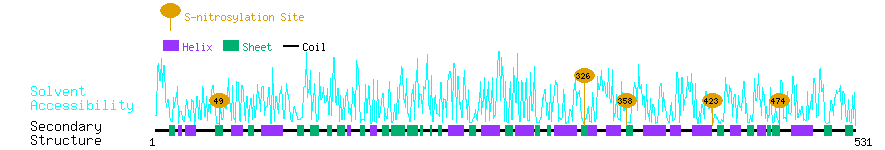 |
|

