Protein Name: E3 ubiquitin-protein ligase Mdm2
|
UniprotKB/SwissProt ID: MDM2_HUMAN (Q00987)
Gene Name: MDM2
Organism: Homo sapiens (Human).
Function: E3 ubiquitin-protein ligase that mediates ubiquitination of p53/TP53, leading to its degradation by the proteasome. Inhibits p53/TP53- and p73/TP73-mediated cell cycle arrest and apoptosis by binding its transcriptional activation domain. Also acts as a ubiquitin ligase E3 toward itself and ARRB1. Permits the nuclear export of p53/TP53. Promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma RB1 protein. Inhibits DAXX-mediated apoptosis by inducing its ubiquitination and degradation. Component of the TRIM28/KAP1-MDM2-p53/TP53 complex involved in stabilizing p53/TP53. Also component of the TRIM28/KAP1-ERBB4-MDM2 complex which links growth factor and DNA damage response pathways. Mediates ubiquitination and subsequent proteasome degradation of DYRK2 in nucleus. Ubiquitinates IGF1R and SNAI1 and promotes them to proteasomal degradation.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Nucleus, nucleoplasm. Cytoplasm. Nucleus, nucleolus. Note=Expressed predominantly in the nucleoplasm. Interaction with ARF(P14) results in the localization of both proteins to the nucleolus. The nucleolar localization signals in both ARF(P14) and MDM2 ma
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | HPRD | 01272 | Accelerated tumor formation, susceptibility to |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 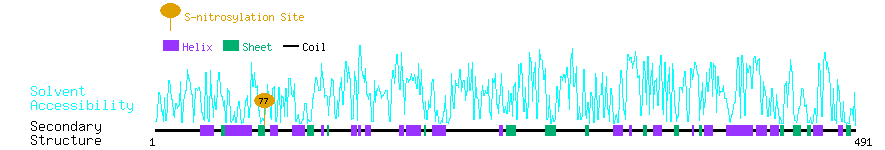 |
|
