Protein Name: Macrophage migration inhibitory factor
|
UniprotKB/SwissProt ID: MIF_HUMAN (P14174)
Gene Name: MIF
Synonyms: GLIF, MMIF
Organism: Homo sapiens (Human).
Function: Pro-inflammatory cytokine. Involved in the innate immune response to bacterial pathogens. The expression of MIF at sites of inflammation suggests a role as mediator in regulating the function of macrophages in host defense. Counteracts the anti- inflammatory activity of glucocorticoids. Has phenylpyruvate tautomerase and dopachrome tautomerase activity (in vitro), but the physiological substrate is not known. It is not clear whether the tautomerase activity has any physiological relevance, and whether it is important for cytokine activity.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Secreted. Cytoplasm. Note=Does not have a cleavable signal sequence and is secreted via a specialized, non- classical pathway. Secreted by macrophages upon stimulation by bacterial lipopolysaccharide (LPS), or by M.tuberculosis antigens.
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | HPRD | 01091 | Rheumatoid arthritis, systemic juvenile, susceptibility to |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 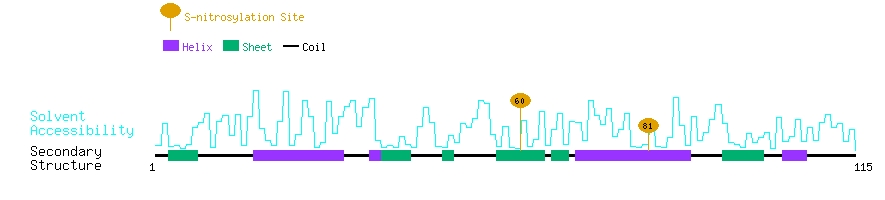 |
|
