Protein Name: Myosin-binding protein C, cardiac-type
|
UniprotKB/SwissProt ID: MYPC3_MOUSE (O70468)
Gene Name: Mybpc3
Organism: Mus musculus (Mouse).
Function: Thick filament-associated protein located in the crossbridge region of vertebrate striated muscle a bands. In vitro it binds MHC, F-actin and native thin filaments, and modifies the activity of actin-activated myosin ATPase. It may modulate muscle contraction or may play a more structural role.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization:
| |
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 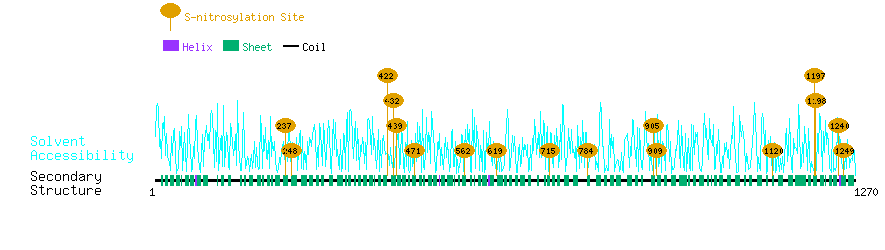 |
|
