Protein Name: Neurofascin
|
UniprotKB/SwissProt ID: NFASC_MOUSE (Q810U3)
Gene Name: Nfasc
Organism: Mus musculus (Mouse).
Function: Cell adhesion, ankyrin-binding protein which may be involved in neurite extension, axonal guidance, synaptogenesis, myelination and neuron-glial cell interactions (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cell membrane; Single-pass type I membrane protein.
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 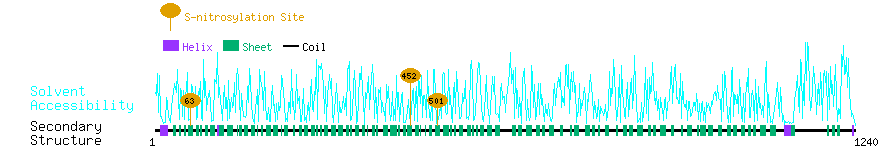 |
|

