Protein Name: Glutamate receptor ionotropic, NMDA 1
|
UniprotKB/SwissProt ID: NMDZ1_HUMAN (Q05586)
Gene Name: GRIN1
Synonyms: NMDAR1
Organism: Homo sapiens (Human).
Function: NMDA receptor subtype of glutamate-gated ion channels with high calcium permeability and voltage-dependent sensitivity to magnesium. Mediated by glycine. This protein plays a key role in synaptic plasticity, synaptogenesis, excitotoxicity, memory acquisition and learning. It mediates neuronal functions in glutamate neurotransmission. Is involved in the cell surface targeting of NMDA receptors (By similarity).
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Cell membrane; Multi-pass membrane protein (By similarity). Cell junction, synapse, postsynaptic cell membrane (By similarity). Cell junction, synapse, postsynaptic cell membrane, postsynaptic density (By similarity). Note=Enriched in postsynaptic plasma
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | OMIM | 138249 | GLUTAMATE RECEPTOR, IONOTROPIC, N-METHYL-D-ASPARTATE, SUBUNIT 1; GRIN1 ;;N-METHYL-D-ASPARTAT | | OMIM | 614254 | MENTAL RETARDATION, AUTOSOMAL DOMINANT 8; MRD8 |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 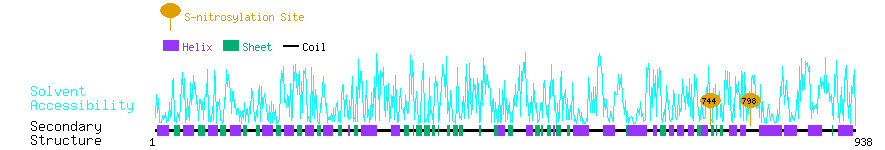 |
|
