Protein Name: Nicotinamide mononucleotide adenylyltransferase 3
|
UniprotKB/SwissProt ID: NMNA3_MOUSE (Q99JR6)
Gene Name: Nmnat3
Organism: Mus musculus (Mouse).
Function: Catalyzes the formation of NAD(+) from nicotinamide mononucleotide (NMN) and ATP. Can also use the deamidated form; nicotinic acid mononucleotide (NaMN) as substrate with the same efficiency. Can use triazofurin monophosphate (TrMP) as substrate. Can also use GTP and ITP as nucleotide donors. Also catalyzes the reverse reaction, i.e. the pyrophosphorolytic cleavage of NAD(+). For the pyrophosphorolytic activity, can use NAD (+), NADH, NAAD, nicotinic acid adenine dinucleotide phosphate (NHD), nicotinamide guanine dinucleotide (NGD) as substrates. Fails to cleave phosphorylated dinucleotides NADP(+), NADPH and NAADP(+). Protects against axonal degeneration following injury.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Mitochondrion.
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 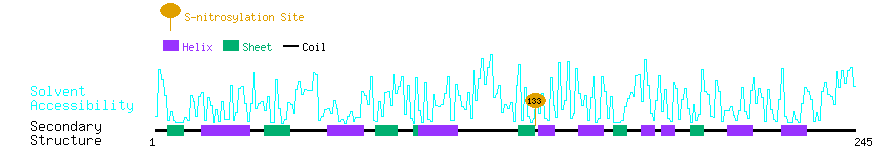 |
|

