Protein Name: Nuclear pore complex protein Nup107
|
UniprotKB/SwissProt ID: NU107_HUMAN (P57740)
Gene Name: NUP107
Organism: Homo sapiens (Human).
Function: Plays a role in the nuclear pore complex (NPC) assembly and/or maintenance. Required for the assembly of peripheral proteins into the NPC. May anchor NUP62 to the NPC.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization: Nucleus membrane. Nucleus, nuclear pore complex. Chromosome, centromere, kinetochore. Note=Located on both the cytoplasmic and nuclear sides of the NPC core structure. During mitosis, localizes to the kinetochores. Dissociates from the dissasembled NPC s
|
PDB :
( If your security settings prevent Jmol from running, please register http://140.138.144.145/ as a safe location in your Java settings. ) |
|
Protein disease:
|
| Disease database |
Database Entry |
Disease information | | OMIM | 607617 | NUCLEOPORIN, 107-KD; NUP107 ;;NUP84, YEAST, HOMOLOG OF; NUP84 |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 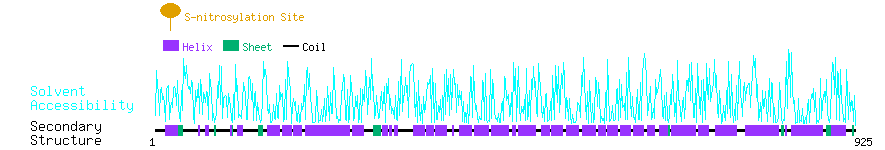 |
|

