Protein Name: ADP-sugar pyrophosphatase
|
UniprotKB/SwissProt ID: NUDT5_MOUSE (Q9JKX6)
Gene Name: Nudt5
Organism: Mus musculus (Mouse).
Function: Hydrolyzes with similar activities ADP-ribose and ADP- mannose. Can also hydrolyzes ADP-glucose (40% of ADP-ribose activity) and other nucleotide sugars with low activity (10%). Does not play a role in U8 snoRNA decapping activity. Binds U8 snoRNA.
Other Modifications: View all modification sites in dbPTM
Protein Subcellular Localization:
| |
|
Network with metabolic pathway:
|
|
Graphical Visualization of S-nitrosylation Sites:
|
| Overview of Protein S-nitrosylation Sites with Functional and Structural Information | 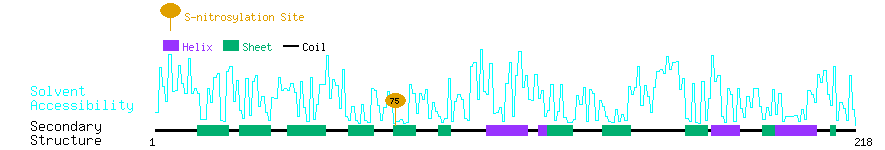 |
|

